Over the last seven years, we have tailored our technology towards challenging target families, including G-protein coupled receptors (GPCRs), Ion Channels, and other transmembrane proteins. These target families have the potential to significantly improve patients' lives.
We and many other drug developers believe this arises from industry focusing on the easier-to-drug targets, as they provide safer and more streamlined revenue generators. This has left researchers to grapple with a panel of more complex diseases needing treatments.
Only 3 FDA-approved antibodies exist for GPCRs despite associations with over 60 diseases. This represents a fraction of the 80+ antibodies available in the market.
There is a clear need to begin focusing our efforts towards challenging targets.
This article will explain our discovery approach to challenging drug targets. In future editions, we will explore each successive step of our discovery process in greater detail.
Without further ado, here is how we discover antibodies for challenging targets:
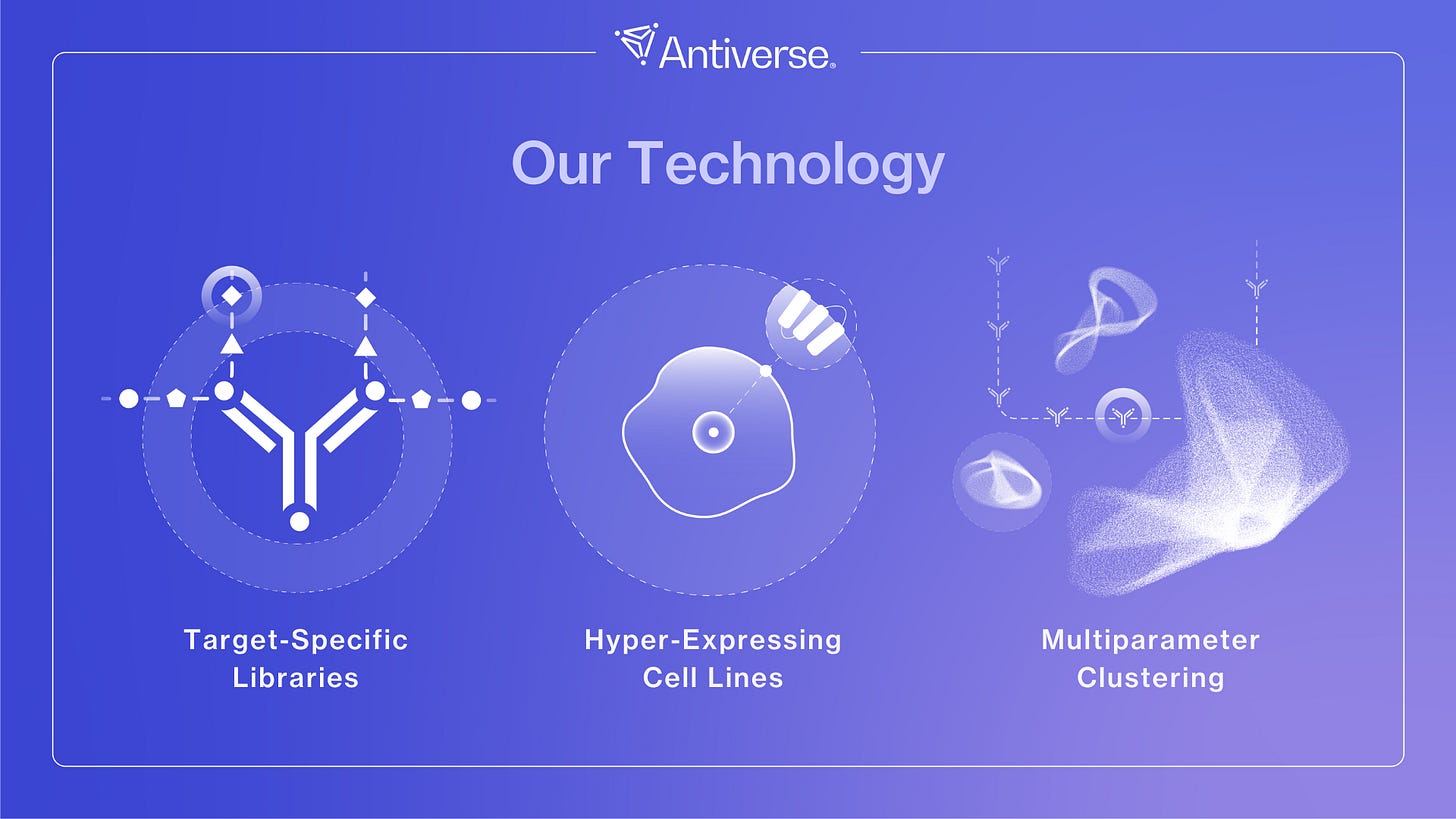
The Major Components Of Our Design Platform
Our AI antibody design platform has three core components: Target-Specific Library Design, Screening via our Proprietary Hyper-Expressing Cell Lines, and the Multiparameter Clustering Module. Let's unpack these in more depth:
1. Target-Specific Library Design.
Traditional antibody libraries contain millions of random binders. In modern antibody discovery methods, these libraries are synthesised and screened against the target of interest to see if one of them binds.
However, binders are extremely rare for challenging targets. Therefore, random sampling approaches typically prove inefficient for antibody discovery against these targets.
Experts estimate that around 227, or 80% of GPCRs with known disease links remain undrugged.
Target-specific libraries are generated using machine learning and designed with high confidence against the desired target. Screening higher-confidence candidates significantly enhances our predictive accuracy, boosting the likelihood of finding effective binders.
2. Proprietary Hyper-Expressing Cell Lines
We convert our target-specific libraries into scFv phage-display libraries.
These libraries are screened against our proprietary cell lines, which host over 1 million receptors per cell.
Typical cell lines struggle to express GPCRs in high densities. A lower number of receptors on the cell surface reduces the sensitivity of the screening process.
Cell lines with high densities of the desired receptor and low densities of other interfering receptors ensure that antibodies are rarely missed during screening.
Combining this with our high-confidence target-specific libraries significantly improves our odds of discovering rare GPCR antibodies.
3. Multiparameter Clustering Module
Positive binding fragments are converted back into antibodies and sequenced.
We feed the sequences to our Multiparameter Clustering Module, an artificial intelligence algorithm that analyses antibody DNA sequences across 20 complex patterns and properties, including similarities not apparent to human observers.
We select the clusters with the best-predicted properties for further functional testing.
Our technology is already helping three global pharmaceutical companies discover antibodies for challenging targets. Combined with our own internal assets, we have 9 active antibody discovery programmes for challenging targets.
You can contact us via our website if you have a target in mind or want to learn more about our technology. We also have a PDF with more details on our technology available for download.
We enable you to work with challenging targets, accelerating your discovery pipeline.
This Month in AI Drug Discovery
EvolutionaryScale has unveiled ESM3, a cutting-edge AI protein language model, alongside securing $142 million in funding. This model, trained on over 2.7 billion protein sequences, aims to innovate drug development and sustainability. It has already created new fluorescent proteins, highlighting its potential to transform medicine and biotechnology.
Link: https://www.nature.com/articles/d41586-024-02214-x / https://www.evolutionaryscale.ai/blog/esm3-release
Iambic Therapeutics has raised $50 million in an oversubscribed Series B+ round led by Mubadala Capital and Exor Ventures. The funds will enhance their AI-driven platform for faster drug discovery and development, integrating AI with high-throughput experimentation to identify promising drug candidates quickly.
Link: https://equalocean.com/news/2024062521050
Ipsen has inked a $1 billion deal with Foreseen Biotechnology for exclusive rights to develop and commercialise FS001, a preclinical ADC targeting solid tumours. Ipsen will prepare FS001 for phase 1 trials, enhancing its oncology pipeline.
Link: https://www.fiercebiotech.com/biotech/ipsen-scores-second-adc-1b-biobucks-pact-foreseen
Researchers have shown that a protein language model, enhanced with structural backbone coordinates, can effectively guide protein evolution. This approach was used to improve the neutralisation and affinity of clinical antibodies against SARS-CoV-2 variants by up to 25-fold and 37-fold, respectively. The study demonstrates the model's potential in protein engineering without requiring task-specific training data.
Link: https://www.fiercebiotech.com/biotech/ipsen-scores-second-adc-1b-biobucks-pact-foreseen
About Antiverse
Antiverse is an artificial intelligence-driven techbio company that specialises in antibody design against challenging targets, including G-protein coupled receptors (GPCRs) and ion channels. Headquartered in Cardiff, UK and with offices in Boston, MA, Antiverse combines state-of-the-art machine learning techniques and advanced cell line engineering to develop de novo antibody therapeutics. With a main focus on establishing long-term partnerships, Antiverse has collaborated with two top 20 global pharmaceutical companies. In addition, they are developing a strong internal pipeline of antibodies against several challenging drug targets across various indications. For more information, please visit
https://www.antiverse.io
Found this article interesting? Share it with your network.